Background:
Somatic mutations (mut) and acute myeloid leukemia (AML) differentiation state (phenotype) influence sensitivity and response to treatment. However the interaction between genotype and phenotype with respect to sensitivity or resistance to venetoclax (VEN) based therapy has yet to be fully evaluated.
Methods:
Samples from patients (pts.) with newly diagnosed (ND) AML enrolled in Beat AML with available bulk RNA sequencing (RNAseq), whole exome sequencing, and ex vivo drug sensitivity testing (N=255) were analyzed for correlation between AML phenotype (defined using RNAseq [van Galen et. al. Cell 2019] and surface immunophenotype via multiparameter flow cytometry [MFC]), genotype, and VEN sensitivity. A monocytic score corresponding to monocytic gene expression and myeloid differentiation for each sample was included as previously defined (Bottomly et. al. Cancer Cell 2022).
Clinical outcomes were assessed in an independent retrospective patient cohort (N=97) treated with frontline hypomethylating agents (HMA)+VEN at Oregon Health & Science University. Categorical and continuous variables were analyzed using Fisher's or Wilcoxon rank sum testing. Time to event outcomes utilized the log-rank method with cox multivariable regression modeling.
Results:
Median patient (pt) age at diagnosis in the surveyed cohort was 61 years (range 20-88). AML phenotype determined using proportional deconvolution of RNAseq data was HSC/progenitor (primitive) in 31% (N=78), mixed/intermediate in 31% (N=79), cDC/monocytic (mature) in 29% (N=73), and unknown in 10% (N=25) of pts.
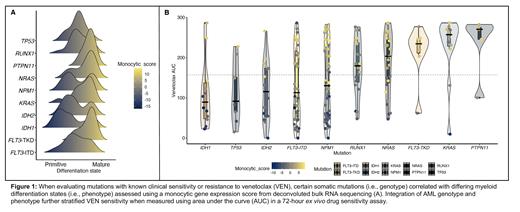
Muts frequent in primitive vs. mature AML samples included transcription factor ( RUNX1, CEBPA, GATA2, GATA1, ETV6, IKZF1; 30% vs. 15%, p=0.050), tumor suppressor ( TP53, PHF6, WT1; 21% vs. 7%, p=0.019), and IDH1/2 (27% vs. 12%, p= 0.027). Active signaling mut ( FLT3-ITD/TKD, K/NRAS, PTPN11, BRAF) were more frequent in mature vs. primitive samples (78% vs. 44%, p < 0.0001), underscoring the correlation between genotype and myeloid differentiation (Fig.A).
Expression of anti-apoptotic protein and myeloid differentiation markers measured using RNAseq varied by AML genotype and phenotype and corresponded to VEN resistance. The ratios of BCL2A1 (pearson R=0.62, p < 0.0001), CD11b (R=0.65, p < 0.0001), CD14 (R=0.73, p < 0.0001), and CLEC7A (R=0.72, p < 0.0001) to BCL2 positively correlated with VEN resistance (i.e., increasing area under the curve [AUC]).
While IDH1/2 and NPM1 mutated samples demonstrated the highest BCL2 expression and lowest BCL2A1: BCL2 ratio and remained largely sensitive to VEN, myeloid differentiation (in particular monocytic differentiation) increased VEN resistance irrespective of genotype. For instance, classification of a primitive vs. mature phenotype further stratified ex vivo VEN sensitivity in pts with IDH1 (median AUC: 61 vs. 195, p: 0.004), IDH2 (median AUC: 61 vs. 185, p: 0.002), or NPM1 (median AUC: 60 vs. 220, p < 0.0001) mut (Fig.B).
Overall survival (OS) following HMA+VEN was evaluated based on the presence or absence of surface immunophenotypic markers measured via MFC that correlated with myeloid differentiation. CD117+ AML (HR: 0.30 95% CI: 0.14 - 0.65, p=0.002) associated with improved OS, while inferior OS was observed with CD11b+ (HR: 2.03 95% CI: 1.07 - 3.86, p=0.031) and CD64+ AML (HR:1.86 95% CI: 0.90 - 3.85, p=0.095).
When assessing the combined influence of genotype and immunophenotype on OS in CD11b+ or CD64+ AML, IDH1/2 mut (N=8) numerically improved OS compared to IDH1/2 wild-type cases (N=17) (median 12.3 vs. 5.8 months, p: 0.17). In multivariate analysis adjusted for CD11b or CD64 positivity, age, IDH1/2 mut, and secondary or therapy-related AML, CD11b or CD64 positivity (HR: 1.98 95% CI: 1.08-3.66, p=0.028) retained the strongest impact on OS.
Conclusions:
Genotype and phenotype both modulate VEN sensitivity in AML. Myeloid differentiation is associated with increased alternative anti-apoptotic protein expression and VEN resistance irrespective of genotype; however certain mutations (i.e., IDH1/2) may retain prognostic significance independent of myeloid differentiation. Given both factors influence VEN sensitivity, prognostic models incorporating AML phenotype and genotype may further improve risk stratification and predict response to VEN based therapy in AML.
Disclosures
Lachowiez:Rigel Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees; COTA Healthcare: Consultancy. Gandhi:orca bio: Research Funding. Leonard:Adaptive Biotechnologies: Consultancy, Membership on an entity's Board of Directors or advisory committees, Other: Travel, accommodations, expenses; Pfizer: Consultancy; Takeda: Consultancy; Kite/Gilead: Consultancy. Maziarz:Gamida: Research Funding; Athersys: Other: Patent holder; Orca Therapeutics: Research Funding; Kite: Consultancy; AlloVir: Consultancy, Research Funding; Novartis: Consultancy, Research Funding. Saultz:IKENA Oncology: Research Funding; Rigel: Other: Advisory Board. Traer:Abbvie: Consultancy, Membership on an entity's Board of Directors or advisory committees; Prelude Therapeutics: Research Funding; Servier: Membership on an entity's Board of Directors or advisory committees; Schrodinger: Research Funding; Incyte: Research Funding; Astra-Zeneca: Research Funding; Astellas: Consultancy, Membership on an entity's Board of Directors or advisory committees; Daiichi-Sankyo: Membership on an entity's Board of Directors or advisory committees; Rigel: Membership on an entity's Board of Directors or advisory committees. Braun:Oryzon Genomics: Other: Institutional PI (FRIDA trial); Novartis: Consultancy; Gilead Sciences: Research Funding; Blueprint Medicines: Consultancy, Research Funding; AstraZeneca: Research Funding. Swords:Kronos Bio: Research Funding. Tyner:Petra: Research Funding; Schrodinger: Research Funding; Tolero: Research Funding; Recludix Pharma: Membership on an entity's Board of Directors or advisory committees; Acerta: Research Funding; Aptose: Research Funding; AstraZeneca: Research Funding; Constellation: Research Funding; Incyte: Research Funding; Genentech: Research Funding; Meryx: Research Funding; Kronos: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal